Aminopeptidase
Aminopeptidases (APs) are a family
of widely distributed proteases which participate in many significant biological
processes, such as protein maturation, hormone production, and peptide digestion
[1]. While several Zn peptidases are known to contain a single
Zn2+ ion in their active site [2], a few
metallo-APs, including those from bovine lens (bAP) [3], Escherichia
coli (eAP) [4], Aeromonas proteolytica (aAP) [5],
and Streptomyces griseus (sAP) [6] have been proved by means of X-ray
crystallography to contain a dinuclear metal active site. However,
diverse structural features and mechanistic properties have been revealed
in these dinuclear metallo-APs [7]; including their different tertiary
and quaternary structures, the lack of a conserved geometry and coordinated
ligand type in the active site, and the observation of dinuclear catalysis
in some APs while mononuclear catalysis in others.
Despite the presence of a dinuclear
site in aAP[5] and presumably in porcine kidney AP (pAP) [8], these
two APs have been observed to exhibit a "mononuclear" catalysis. These
two enzymes have been previously shown to exhibit a selective metal binding
property, where a full hydrolytic activity was detected when one metal ion
was bound [8,9] A modulation of the activity was observed when
a second metal ion was introduced, indicating a regulatory role [10].
Nevertheless, this mononuclear catalysis was not observed in bAP action [3].
Further studies of other dinuclear APs are therefore necessary to provide
more mechanistic information about this mononuclear and dinuclear discrepancy
in AP action.
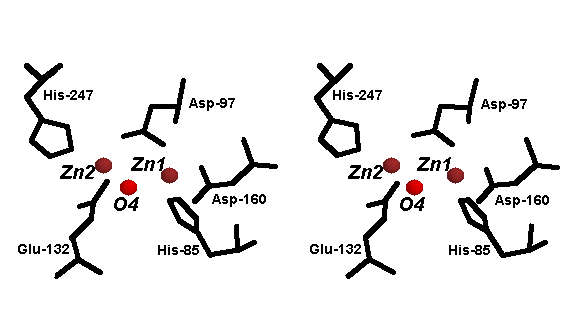
The AP isolated from the culture
medium of Streptomyces griseus (sAP, MW ~30 kDa) has been characterized to
contain 2 Zn2+ ions per molecule (see structure, and the active site structure shown
below) [11]. A previous metal-activation study using the slow
substrate Ala-p-nitroanilide revealed that Mn2+,
Co2+, or Zn2+
was bound simultaneously to the two metal binding sites of the enzyme at
pH 8 in the presence of 1 mM Ca2+; i.e.,
the activity of the enzyme was parallel to the amount of metal ion bound
to the enzyme and reached a plateau with 2 equivalent metal ions introduced
[11a]. Since the two metal sites could not be selectively filled with
metal ions in the previous study, identification of each metal site was not
possible and the role of each metal ion in catalysis could not be easily
revealed. It is important to find out the conditions for a selective
metal binding to the metal sites, which is an inevitable step to provide
structural information and catalytic role about each individual metal site
by means of physical methods.
 We have been studying the metal
binding properties, hydrolytic activity, and active site structure of sAP
by means of activity assay and spectroscopic methods. A sequential Co2+ binding to this enzyme in MES buffer at pH 6.1
has been concluded on the basis of optical study, isotropically shifted 1H NMR features, and activity assay. Moreover,
we have shown a very rare case that Cu2+
ion, which is better known as an NMR "relaxation probe", can be utilized as
a "shift probe" for the study of metal binding sites in metalloproteins and
affords sharp hyperfine-shifted 1H NMR
signals (see abstract).
The presence of a dinuclear metal active site in sAP has been established
by means of 1H NMR using Cu2+ as a probe. This study also demonstrates
that although sAP and aAP have nearly identical active sites on the basis
of their crystal structures [5,6] (however, with a low sequence homology
~30% [12]), they exhibit quite different mechanisms in that sAP shows a dinuclear
hydrolytic catalysis whereas aAP shows a mononuclear catalysis.
We have been studying the metal
binding properties, hydrolytic activity, and active site structure of sAP
by means of activity assay and spectroscopic methods. A sequential Co2+ binding to this enzyme in MES buffer at pH 6.1
has been concluded on the basis of optical study, isotropically shifted 1H NMR features, and activity assay. Moreover,
we have shown a very rare case that Cu2+
ion, which is better known as an NMR "relaxation probe", can be utilized as
a "shift probe" for the study of metal binding sites in metalloproteins and
affords sharp hyperfine-shifted 1H NMR
signals (see abstract).
The presence of a dinuclear metal active site in sAP has been established
by means of 1H NMR using Cu2+ as a probe. This study also demonstrates
that although sAP and aAP have nearly identical active sites on the basis
of their crystal structures [5,6] (however, with a low sequence homology
~30% [12]), they exhibit quite different mechanisms in that sAP shows a dinuclear
hydrolytic catalysis whereas aAP shows a mononuclear catalysis.
Our publications about Streptomyces aminopeptidase:
- Lin, L.-Y.; Park, H. I.; Ming, L.-J.* "Metal Binding and Active Site
Structure of Di-Zinc Streptomyces griseus Aminopeptidase" J. Biol.
Inorg. Chem. 1997, 2, 744-749. (abstract
and reprint in PDF)
- Holz, R. C.*; Bennett, B.; Chen, G.; Ming, L.-J. "Proton NMR Spectroscopy
as a Probe of Dinuclear Copper(II) Active Sites in Metalloproteins.
Charaterization of the Hyperactive Copper(II)-Substituted Aminopeptidase from
Aeromonas proteolytica" J. Am. Chem. Soc. 1998, 120,
6329-6335. (abstract
and reprint in PDF )
- Ming, L.-J. "NMR Studies of Paramagnetic Multinuclear Metalloproteins"
Trends in Inorganic Chemistry 1998, 5, 205–236. (Table
of contents and Introduction)
- Harris, M. N.; Ming, L.-J.* "Different Phosphate Binding Modes of
Streptomyces griseus Aminopeptidase between Crystal and Solution
States and the Status of Zinc-Bound Water" FEBS Lett. 1999,
455, 321-324. (abstract
and reprint in PDF)
- Park, H. I.; Ming, L.-J.* "A 1010
Rate Enhancement of Phosphodiester Hydrolysis by a Di-Zinc Aminopeptidase–
Transition State Analogues as Substrates?" Angew. Chem. Intl. Engl. Ed.
1999, 38, 2914-2916. (abstract
and reprint in PDF )
- Ercan, A.; Park, H. I.; Ming, L.-J.* "Remarkable Enhancement of the
Hydrolyses of Phosphoesters by Dimetal Centers —Streptomyces Aminopeptidase
as a “Natural Model System”" Chem. Commun. 2000, 2501–2502.
(abstract
and reprint)
- Hasselgren, C.; Park, H. I.; Ming, L.-J.* "Metal Ion Binding and Activation
of Streptomyces griseus Aminopeptidase —Cadmium(II) Binding as a Model"
J. Biol. Inorg. Chem. 2001, 6, 120–127. (abstract
and reprint)
- Harris, M. N.; Bertolocci, C.; Ming, L.-J.* "31P
NMR Relaxation and Kinetic Studies of Cobalt(II)-Substituted Streptomyces
Dinuclear Aminopeptidase" Inorg. Chem. 2002, 41, 5582-5588. (abstract
and reprint)
References
- (a) Taylor A (1993) FASEB J 7:290-298.
(b) Taylor A (1993) TIBS 18:167-172. (c) Gonzales T, Robert-Baudouy
J (1996) FEMS Microbiol Rev 18:319-344.
- (a) Christianson DW, Lipscomb WN (1989) Acc
Chem Res 22:62-69. (b) Mangani S, Carloni P, Orioli P (1992) Coord
Chem Rev 120:309-324. (c) Matthews BW (1988) Acc Chem Res
21:333-340.
- (a) Burley SK, David PR, Taylor A, Lipscomb
WN (1990) Proc Natl Acad Sci USA 87:6878-6882. (b) Burley SK,
David P R, Sweet R M, Taylor A, Lipscomb WN (1992) J Mol Biol 224:113-140.
(c) Sträter N, Lipscomb WN (1995) Biochemistry 34:9200-9210.
(d) Kim H, Lipscomb WN (1994) Adv Enzymol 68:153-213.
- Roderick SL, Matthews BW (1993) Biochemistry
32,:907-3912.
- Chevrier B, Schalk C, D'Orchymont H, Rondeau
J-M, Moras D, Tarnus C (1994) Structure 2:283-291.
- Greenblatt HM, Almog O, Maras B, Spungin-Bialik
A, Barra D, Blumberg S, Shoham G (1997) J Mol Biol 265:620-636.
- (a) Sträter N, Lipscomb WN, Klabunde T,
Krebs B (1996) Angew Chem Int Ed Engl 35:2024-2055. (b) Lipscomb
WN, Sträter N (1996) Chem Rev 96:2375-2433.
- Van Wart HE, Lin SH (1981) Biochemistry
20:5682-5689.
- Prescott JM, Wagner FW, Holmquist B, Vallee
BL (1985) Biochemistry 24:5350-5356.
- (a) Vallee BL, Auld DS (1993) Proc Natl Acad
Sci USA 90:2715-2718. (b) Vallee BL, Auld DS (1993) Biochemistry
32:6493-6500.
- (a) Ben-Meir D, Spungin A, Ashkenazi R, Blumberg
S (1993) Eur J Biochem 212:107-112. (b) Spungin A, Blumberg S
(1989) Eur J Biochem 183:471-477.
- Maras B, Greenblatt HM, Shoham G, Spungin-Bialik
A, Blumberg S, Barra D (1996) Eur J Biochem 236:843-846.

